|
|
|
|
| The Chemist Volume 91 | Number 1 |
 
|
|
| |
|
Using Protein Function Prediction to Promote
Hypothesis-Driven Thinking in
Undergraduate Biochemistry Education
|
| |
Abstract: Students at the Rochester Institute of Technology and Dowling College used bioinformatics software, which they had helped develop, to predict the function of protein structures whose functions had not been assigned or confirmed. Over the course of time, they incorporated other bioinformatics tools and moved the project to the wet lab, where they sought to confirm their in silico predictions with in vitro assays. In this process, we saw so much personal and professional growth among our students that we chose to implement their approach in an undergraduate biochemistry teaching lab, which we call BASIL, for Biochemistry Authentic Scientific Inquiry Lab. This curriculum has now been implemented by thirteen faculty members on eight campuses, and we look forward to a long-range exploration of BASIL’s impact on the students who enroll in courses that use the BASIL curriculum.
Key Words: Biochemistry education, bioinformatics, protein function, structure alignment, course-based undergraduate research experience.
|
|
|
Student Initiative Led to Dramatic Change in our Research
Early in my career at the Rochester Institute of Technology (RIT), [note: The first-person narrative is from the point of view of Paul Craig] a colleague shared his ideas with me about how to design an undergraduate biochemistry laboratory course that would engage the students in a research endeavor. Starting from his ideas, I created a project-based undergraduate biochemistry laboratory course where students purified and studied a single enzyme using various techniques [1]. I hoped to introduce a basic research component to the course with techniques such as site-directed mutagenesis, but was never able to move the students that far along with the project. At the same time, it was my privilege to direct independent research with a steady stream of inquisitive undergraduate students and found that much more rewarding.
In 2004, Herbert Bernstein and I began collaborating on a project funded by the NSF ATE program that was centered on using 3D visualization across the scientific curriculum, where we focused on molecular visualization. We chose to work with the PyMOL molecular graphics environment because it contained a number of 3D visualization modes. Two of the students on that project, both bioinformatics majors, created a simplified user interface for PyMOL called EZ-Viz, which was designed to help educators use PyMOL without having to learn commands in the Python language [2]. The following year, two additional students built ProMOL, based on the original code of EZ-Viz. ProMOL accesses additional tools in PyMOL to enable the user to create motifs of enzyme active sites, then search for those motifs in any protein that has 3D coordinates [3]. Remarkably, these students were biotechnology majors with no background in programming, yet they taught themselves Python as they were developing the code for ProMOL (source code can be found at https://github.com/SBEVSL/sbevsl_migrated/tree/master/promol). These students moved us another step toward our goal of merging the teaching lab with authentic research activities by converting a teaching tool (EZ-Viz) into a research tool (ProMOL).
Students in our research lab then began using ProMOL to predict functions for some of the >3000 proteins in the Protein Data Bank [4,5] that are described as having an “unknown function,” many of which resulted from the Protein Structure Initiative [6]. In one of the first cases we studied in depth, an undergraduate student aligned PDB entry 3DS8 [7] against the library of enzyme active sites found in ProMOL and found an excellent alignment with PDB entry 1ORV, a porcine dipeptidyl peptidase which was also a serine hydrolase [8]. He immediately wanted to confirm his findings in the wet lab, so he obtained the plasmid containing the gene for 3DS8, expressed the protein, purified it, and analyzed it by SDS-PAGE to confirm that the protein was pure and had the expected molecular weight. Initial testing with a QuanticleaveTM protease assay kit (ThermoFisher Scientific) revealed a low level of proteolytic activity. He decided that this was clearly not a protease, so he began looking for other tools to help him make a better prediction that he could test in the lab. The student then analyzed the sequence of 3DS8 using BLAST [9] and found some of the best sequence alignments were with the esterase/lipase superfamily. Further analysis within ProMOL revealed a very good active site alignment between 3DS8 and PDB entry 1TAH (Figure 1), a lipase with a known and well documented active site [10]. He then shifted his emphasis in the wet lab to test activity with colorimetric esterase substrates. In this process, the student had shifted our research from being strictly in silico to becoming in vitro. It was clear that his curiosity had been piqued by the computational discoveries, but that it would only be satisfied by moving to the bench to verify his predictions.
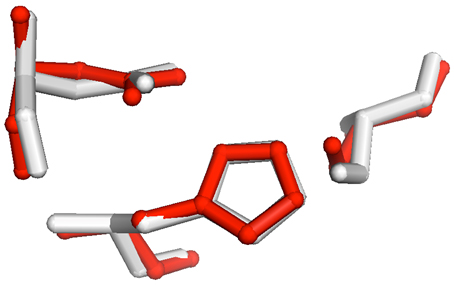
Fig 1. Alignment of PDB entry 3DS8 (red) with triacylglycerol lipase,
PDB entry 1TAH (white).
The RMSD for the all atom alignment for the three active site residues (aspartate 263 in 1TAH/aspartate 188 in 3DS8; histidine 285/histidine 222; serine 87/serine 102) was 0.42 Å . |
After this initial success, the students in our research group realized that they needed to access additional tools to effectively predict protein function based only on the 3D coordinates of protein structures. They subsequently developed the flow chart shown in Figure 2 that integrated results from BLAST [9], Pfam [11], and Dali [12] with our findings with ProMOL. A team of five or six students in our research group then analyzed >3000 protein structures of unknown function from the PDB, which led to >50 promising predictions of protein function [13].
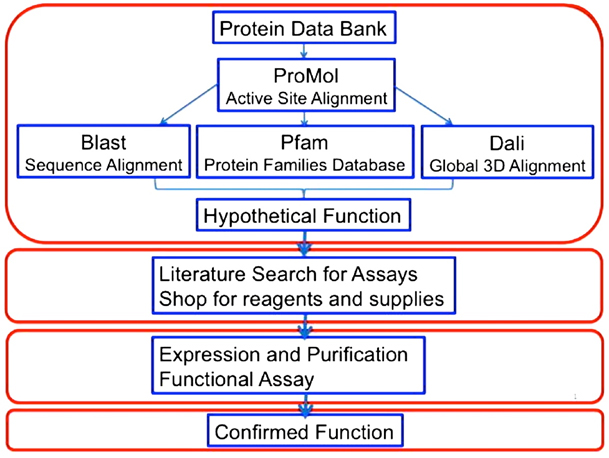
Fig 2. A flowchart for protein function prediction and in vitro confirmation of enzyme activity. This figure was published previously [14] and is used with permission. |
The faculty members leading this project (Herbert J. Bernstein, Jeffrey L. Mills, and Paul A. Craig) noticed a clear pattern throughout this project over the course of a decade; undergraduate students began with our suggestions, but quickly moved beyond our suggestions to start asking new questions, formulating new hypotheses, and trying new things in the computational lab and in the wet lab. In our research lab, we provided the students with research goals and a set of tools to pursue those goals. In repeated cases, they pushed through the limits of our approach and started asking questions, such as:
“What other things can we do with these tools?”
“What other tools are available that we can use?”
“What tools can we create to better pursue our questions?”
This was reinforced as we watched our students present their findings at conferences. They were thoughtful and effective in their presentations and engaged as peers in dialogue with established scientists;
asking and answering questions, discussing shortcomings in their results or their protocols, and seeking and sharing insights into future directions. In one case, a junior undergraduate submitted an abstract to present a poster at the annual meeting of the American Society for Biochemistry & Molecular Biology (ASBMB). Based on her abstract, she was invited to give a talk at the conference. She presented her talk in a session with two established investigators, a post-doc, and a doctoral student. Her presentation was well received and she answered questions accurately and with confidence. Following the session, she asked me why I did not tell her that she would be the only undergraduate presenting a talk in that session. I assured her that I was confident in her knowledge of the project, her understanding of where it fit with the larger scientific questions being addressed in the session, and in her ability to handle herself in a stressful situation. In short, she and the other students engaged in this project were becoming scientists. At that point, we began to consider how we might extend this approach to a larger group of students through an undergraduate biochemistry laboratory course.
Creation of a Course-Based Undergraduate Research Experience
We presented our ideas to our program officer at NIH, who pointed us to the NSF IUSE program. Along with biochemistry colleagues from other universities, we submitted a proposal and received some helpful guidance, mainly that we needed to bring more people on board. Through connections during a poster session at the 2014 ASBMB national conference, we added colleagues with expertise in biochemistry, molecular biology, computational chemistry, and biochemistry education to our team (Table 1), and were able to win funding in the next round.
Table 1. Faculty members and institutions on the project
| Institution |
Faculty Members |
|
| Cal Poly San Luis Obispo |
Anya Goodman, Ashley Ringer McDonald |
| Hope College |
Mike Pikaart |
| Oral Roberts University |
Bob Stewart |
| Purdue University |
Trevor Anderson, Stefan Irby |
| RIT |
Herbert Bernstein, Paul Craig, Jeff Mills, Suzanne O’Handley |
| St. Mary’s University |
Colette Daubner |
| SUNY Oswego |
Julia Koeppe |
| Ursinus College |
Rebecca Roberts |
|
|
|
We are exploring a number of questions as we implement this laboratory experience:
- Can we convert our research lab experience of developing scientists into a Course-based Undergraduate Research Experience (CURE)?
- Can we develop methods to monitor student progress as scientists in a teaching lab?
- How can we, as faculty members, learn to help students grow as scientists?
We faced a number of initial challenges. The first challenge was communication; the team consists of
thirteen faculty members on eight campuses in three different time zones. Our solution was to meet weekly in a video chat room hosted by BlueJeans (bluejeans.com), which is licensed on the RIT campus. Our second challenge was the creation of a uniform set of lab/curriculum modules that were sufficiently flexible to be adapted to courses on campuses with different schedules (semesters vs. quarters, length of lab periods, student availability outside of scheduled lab time), varying instrumentation resources, varying levels of expertise, and comfort in both the wet lab and the computer lab. We created a series of ten modules for the computer lab and the wet lab (Table 2). Creating the modules have been an iterative process over a two-year period (a detailed description of these modules will be the subject of a future manuscript). Our third challenge resulted from our varying levels of expertise, particularly with using computational tools in the lab with our students. During the summer of 2015, we held extended online conversations about the software tools for the project; where we discussed how to install the software, how the software works, and how we can teach our students to use it to make in silico discoveries about protein function. These conversations led members of our team to create a number of tutorials that focus on testing and practicing these computational techniques (basiliuse.blogspot.com).
During our discussions, it became clear that our project fit the description of a CURE [15]. We are aware of several CUREs that focus on nucleic acids, including SEA-PHAGES [16] and the Genomics Education Partnership [17–19], but few that focus on protein structure and function [20]. At that time, we also adopted the acronym BASIL, for Biochemistry Authentic Scientific Inquiry Lab. Current resources for BASIL, including student modules for the lab and video tutorials, can be accessed from the BASIL blog (basiliuse.blogspot.com).
After the first year of the BASIL project, I conducted a survey of the faculty members that focused on their experience of transitioning to a CURE format to teach an undergraduate biochemistry lab. Survey questions were formed by discussions with Trevor Anderson and Stefan Irby, team members from Purdue who are biochemistry educational researchers, and the educational evaluators for the project. The results of the survey have been reported elsewhere [14]. Here is a brief summary of some of the themes that emerged from the survey:
- It would have been better to firmly establish the modules before implementing them on our campuses.
- It is important to communicate regularly, especially when dealing with challenges, while installing or using the software for the project.
- All participants were eager to share their experiences at national conferences of the American Chemical Society, American Biophysical Society, American Society for Biochemistry & Molecular Biology, and the Biennial Conference on Chemical Education.
- All the team members shared enthusiasm for the
project. One stated, “I hope that this research approach will become the norm”.
- All agreed that we need to formally evaluate both the attitudes and the learning experiences of the students involved in the project on our campuses.
Our future plans include creating fully annotated instructor resources for each of the modules, formal assessment of student growth as scientists that results from this course, recruitment of additional campuses to implement the BASIL curriculum, and publication of our students’ findings in the literature on online protein resources.
Table2. Online resources for the BASIL curriculum.
These can be downloaded and viewed at basiliuse.blogspot.com.
| In vitro modules |
In silico modules |
Tutorials for in silico modules |
|
| Protein Expression |
BLAST |
Biochemistry Lab BLAST Tutorial |
| Protein Purification |
Dali |
Biochemistry Lab Dali Tutorial |
| Protein Concentration |
Pfam |
Biochemistry Lab Pfam Tutorial |
| SDS-PAGE |
ProMOL |
Introduction to PyMOL
Introduction to ProMOL
Motif Finder in ProMOL
Structure Query |
| Enzyme Activity |
PyRX |
Ligand Docking with PyRX |
|
|
|
I was invited to present our project at chemistry and biochemistry departments on several campuses during the past year. On two of the campuses, I shortened my presentation to about 25 minutes, and then engaged the audience in a discussion about becoming scientists. The audiences (25-40 people on each campus) were divided into groups of 4-6 people, including graduate students and faculty members. Each group was asked to discuss, “When did you first start to see yourself as a scientist?” Their answers are summarized in Figure 3.
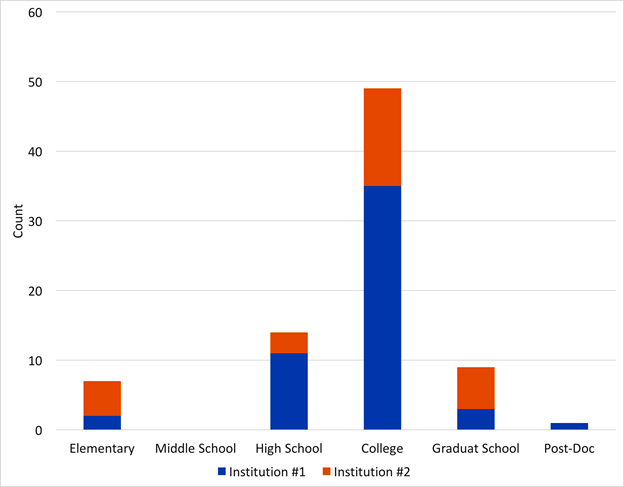
Fig 3. Recollection of seminar participants of their first
self-identification
as scientists for students and faculty on two major campuses. |
The second question for these groups was “How can you tell that students are growing as scientists?”. The groups from these two campuses gave the following responses, which will contribute to our future conversations about student growth as scientists in the BASIL project:
- When “what” questions become “why” questions
- When curiosity becomes more important than grades
- When students start designing their own experiments
- When students challenge the instructor
- When students find a better model on their own
- When students begin to engage in “what if” thinking
- When students start teaching each other
Conclusion
The BASIL project emerged from the minds and actions of a group of undergraduate research students focused on predicting functions for unannotated protein structures. BASIL is a Course-based Undergraduate Research Experience that is led by faculty members on eight different college campuses that include several undergraduate liberal arts institutions, two large public institutions, one large private institution, and a Hispanic-serving institution. The BASIL modules combine wet bench techniques that are common to most undergraduate biochemistry lab courses with bioinformatics skills for the analysis of protein structures and ligand binding to those structures. As a group, we will continue to refine this curriculum so that it can be readily implemented in a wide variety of campus settings. We are also committed to pursuing larger questions associated with this effort:
- Can our CURE offer benefits of research experience to a larger group of students in a classroom setting?
- How do we define the developmental stages of a scientist?
- What are the observable characteristics that demarcate the transition to becoming a scientist?
The authors welcome public comment on these questions on our blog, basiliuse.blogspot.com.
Acknowledgments
I would like to thank the many students who have contributed to this project, with particular gratitude to Brett Hanson, Greg Dodge, and Kaitlin Hart; each dynamically shaped the project and carried our whole research group forward to a new level. This material is based upon work supported by the National Science Foundation under Award #1503811, the National Institute of General Medical Sciences under Award #78077, and the campuses represented in the BASIL project. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation, the National Institute of General Medical Sciences, or the campuses represented in the BASIL project.
References
- Craig PA. A project-oriented biochemistry laboratory course. J. Chem. Educ., 1999, 76, 1130–1135. doi: 10.1021/ed076p1130.
- Grell L, Parkin C, Slatest L, Craig PA. EZ-Viz, a tool for simplifying molecular viewing in PyMOL. Biochem. Mol. Biol. Educ., 2006, 34, 402–407. doi: 10.1002/bmb.2006.494034062672.
- Hanson B, Westin C, Rosa M, Grier A, Osipovitch M, MacDonald ML, Dodge G, Boli PM, Corwin CW, Kessler H, McKay T, Bernstein HJ, Craig PA. Estimation of protein function using template-based alignment of enzyme active sites. BMC Bioinf., 2014, 15, 87. doi: 10.1186/1471-2105-15-87.
- Bernstein FC, Koetzle TF, Williams GJB, E. F. Meyer J, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 1977, 112, 535–542.
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res., 2000, 28, 235–242. doi: 10.1093/nar/28.1.235.
- Berman HM, Westbrook JD, Gabanyi MJ, Tao W, Shah R, Kouranov A, Schwede T, Arnold K, Kiefer F, Bordoli L, Kopp J, Podvinec M, Adams PD, Carter LG, Minor W, Nair R, Baer JL. The protein structure initiative structural genomics knowledgebase. Nucleic Acids Res., 2009, 37, D365–D368. doi: 10.1093/nar/gkn790.
- Zhang R, Wu R, Freeman L, Joachimiak, A. The crystal structure of the gene lin2722 products from Listeria innocua. Be Publ. doi: 10.2210/pdb3ds8/pdb.
- Engel M, Hoffmann T, Wagner L, Wermann M, Heiser U, Kiefersauer R, Huber R, Bode W, Demuth HU, Brandstetter H. The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism. Proc. Natl. Acad. Sci. U.S.A., 2003, 100, 5063–5068. doi: 10.2210/pdb1orv/pdb.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 1997, 25, 3389–3402. doi: 10.1093/nar/25.17.3389.
- Noble ME, Cleasby A, Johnson LN, Egmond MR, Frenken LG. The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate. FEBS Lett., 1993, 331, 123–128. doi: 10.1016/0014-5793(93)80310-Q.
- Finn RD, Miller BL, Clements J, Bateman A. iPfam: a database of protein family and domain interactions found in the Protein Data Bank. Nucleic Acids Res., 2014, 42, D364-373. doi: 10.1093/nar/gkt1210.
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res., 2010, 38, W545–W549. doi: 10.1093/nar/gkq366.
- McKay T, Hart K, Tedla-Boyd W, Horn A, Kessler H, Bernstein HJ, Craig PA. Annotation of Proteins of Unknown Function: Initial Enzyme Results. J Struct Funct Genomics, 2015, 16, 43-54. doi: 10.1007/s10969-015-9194-5.
- Craig PA. A survey on faculty perspectives on the transition to a biochemistry course-based undergraduate research experience laboratory. Biochem. Mol. Biol. Educ., 2017, 45, 426-436. doi: 10.1002/bmb.21060.
- Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M, Trautmann NM, Varma-Nelson P, Weston TJ, Dolan EL. Assessment of Course-Based Undergraduate Research Experiences: A Meeting Report. CBE-Life Sci. Educ., 2014, 13, 29–40. doi: 10.1187/cbe.14-01-0004.
- Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SCR, Findley AM, Gissendanner CR, Golebiewska UP, Guild N, Hartzog GA, Grillo WH, Hollowell GP, Hughes LE, Johnson A, King RA, Lewis LO, Li W, Rosenzweig F, Rubin MR, Saha MS, Sandoz J, Shaffer CD, Taylor B, Temple L, Vazquez E, Ware VC, Barker LP, Bradley KW, Jacobs-Sera D, Pope WH, Russell DA, Cresawn SG, Lopatto D, Bailey CP, Hatfull GF. A Broadly Implementable Research Course in Phage Discovery and Genomics for First-Year Undergraduate Students. mBio, 2014, 5e01051-13. doi: 10.1128/mBio.01051-13.
- Lopatto D, Alvarez C, Barnard D, Chandrasekaran C, Chung H-M, Du C, Eckdahl T, Goodman AL, Hauser C, Jones CJ, Kopp OR, Kuleck GA, McNeil G, Morris R, Myka JL, Nagengast A, Overvoorde PJ, Poet JL, Reed K, Regisford G, Revie D, Rosenwald A, Saville K, Shaw M, Skuse GR, Smith C, Smith M, Spratt M, Stamm J, Thompson JS, Wilson BA, Witkowski C, Youngblom J, Leung W, Shaffer C, Buhler J, Mardis E, Elgin SCR. UNDER GRADUATE RESEARCH. Science, 2008, 322, 684–685. doi: 10.1126/science.1165351.
- Shaffer CD, Alvarez CJ, Bednarski AE, Dunbar D, Goodman AL, Reinke C, Rosenwald AG, Wolyniak MJ, Bailey C, Barnard D, Bazinet C, Beach DL, Bedard JEJ, Bhalla S, Braverman J, Burg M, Chandrasekaran V, Chung H-M, Clase K, DeJong RJ, DiAngelo JR, Du C, Eckdahl TT, Eisler H, Emerson JA, Frary A, Frohlich D, Gosser Y, Govind S, Haberman A, Hark AT, Hauser C, Hoogewerf A, Hoopes LLM, Howell CE, Johnson D, Jones CJ, Kadlec L, Kaehler M, Key SCS, Kleinschmit A, Kokan NP, Kopp O, Kuleck G, Leatherman J, Lopilato J, MacKinnon C, Martinez-Cruzado JC, McNeil G, Mel S, Mistry H, Nagengast A, Overvoorde P, Paetkau DW, Parrish S, Peterson CN, Preuss M, Reed LK, Revie D, Robic S, Roecklein-Canfield J, Rubin MR, Saville K, Schroeder S, Sharif K, Shaw M, Skuse G, Smith CD, Smith MA, Smith ST, Spana E, Spratt M, Sreenivasan A, Stamm J, Szauter P, Thompson JS, Wawersik M, Youngblom J, Zhou L, Mardis ER, Buhler J, Leung W, Lopatto D, Elgin SCR. A Course-Based Research Experience: How Benefits Change with Increased Investment in Instructional Time. CBE-Life Sci. Educ., 2014, 13:111–130. doi: 10.1187/cbe-13-08-0152.
- Lopatto D, Hauser C, Jones CJ, Paetkau D, Chandrasekaran V, Dunbar D, MacKinnon C, Stamm J, Alvarez C, Barnard D, Bedard JEJ, Bednarski AE, Bhalla S, Braverman JM, Burg M, Chung H-M, DeJong RJ, DiAngelo JR, Du C, Eckdahl TT, Emerson J, Frary A, Frohlich D, Goodman AL, Gosser Y, Govind S, Haberman A, Hark AT, Hoogewerf A, Johnson D, Kadlec L, Kaehler M, Key SCS, Kokan NP, Kopp OR, Kuleck GA, Lopilato J, Martinez-Cruzado JC, McNeil G, Mel S, Nagengast A, Overvoorde PJ, Parrish S, Preuss ML, Reed LD, Regisford EG, Revie D, Robic S, Roecklien-Canfield JA, Rosenwald AG, Rubin MR, Saville K, Schroeder S, Sharif KA, Shaw M, Skuse G, Smith CD, Smith M, Smith ST, Spana EP, Spratt M, Sreenivasan A, Thompson JS, Wawersik M, Wolyniak MJ, Youngblom J, Zhou L, Buhler J, Mardis E, Leung W, Shaffer CD, Threlfall J, Elgin SCR. A Central Support System Can Facilitate Implementation and Sustainability of a Classroom-Based Undergraduate Research Experience (CURE) in Genomics. CBE-Life Sci. Educ., 2014, 13, 711–723. doi: 10.1187/cbe.13-10-0200.
- Gray C, Price CW, Lee CT, Dewald AH, Cline MA, McAnany CE, Columbus L, Mura C. Known structure, unknown function: An inquiry-based undergraduate biochemistry laboratory course. Biochem. Mol. Biol. Educ., 2015, 43, 245–262. doi: 10.1002/bmb.20873.
|



